How does astrovirus VA1 cause encephalitis?
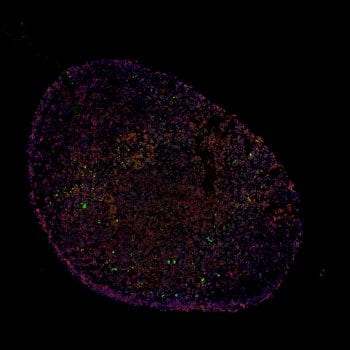
Astrovirus VA1 is the most frequently identified cause of astrovirus-associated encephalitis. We were the first to have successfully isolated and propagated this virus in cell culture. We have since developed the first cell culture system to study the pathogenesis of infection in the central nervous system, as VA1 can infect astrocytes in cell culture. Surprisingly, the virus does not infect neurons, despite the identification of neuronal injury and death from the cases of VA1 encephalitis. These findings to suggest the possibility of VA1 infection of astrocytes leads to a signaling pathway that causes neuronal death. One potential mechanism is through CXCL10 signaling. CXCL10 is induced by VA1 infection and can trigger neuronal apoptosis in a lymphocyte independent mechanism. We are now characterizing these paracellular pathways through a brain organoid model of infection.
Can we develop an animal model of VA1 infection?
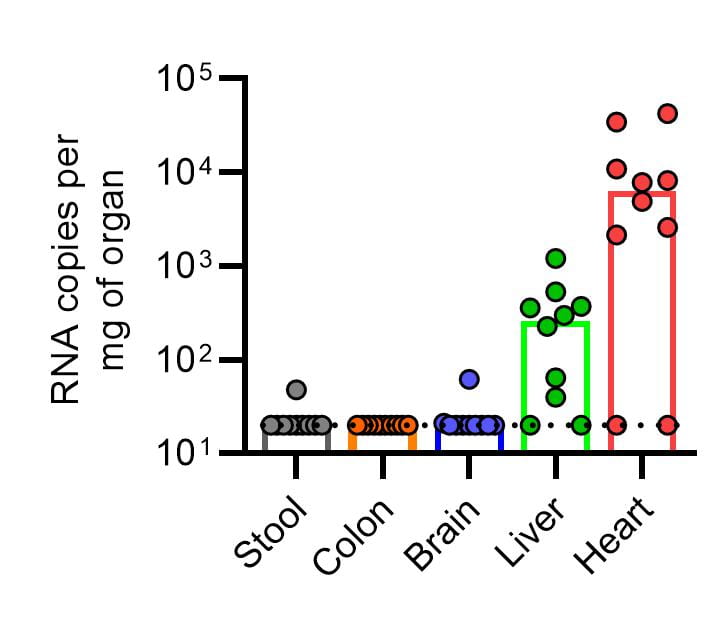
One of the major limitations to the study of astroviruses is the lack of animal models to study astroviruses that infect humans. A turkey model of astrovirus infection exists, but has limited tools. In addition, a murine astrovirus infects mice, but causes no apparent disease. We have used our VA1 isolate and inoculated mice with the hypothesis that it would cause encephalitis. Surprisingly, we do not identify the virus in brain tissue, rather the virus localizes to the heart and is associated with myocarditis. This suggests the potential for astroviruses to cause cardiovascular diseases in humans. We are now studying how the virus causes cardiac injury and the cell types that support infection.
Can we use astrovirus VA1 to study fundamental biology that translates to other viruses?

The stem-loop II motif (s2m) is an RNA sequence present in astroviruses, coronaviruses, picornaviruses, caliciviruses, and reoviruses. Despite the identification of the s2m over 25 years ago, the role of the s2m for the viral lifecycle is poorly understood. We have developed a reverse genetics system for astrovirus VA1 and a partial deletion of the s2m sequence results in recombinant virus that cannot be propagated in cell culture. This phenotypes can be recapitulated if we create a single point mutation in the s2m at nucleotide positions that form guanine-cytosine (G-C) hydrogen base pairs. The virus can be rescued when we create a compensatory substitution that restores the G-C base pair, but is inverted relative to the wild-type s2m sequence. We are now dissecting the mechanism by which the s2m is essential for the astrovirus lifecycle. In addition, we have found the s2m to be important for human astrovirus 1, but surprisingly, the s2m is dispensable for SARS-CoV-2. Once we understand the function of the s2m, we can elucidate why the s2m has different functional roles for the viral lifecycle.

Electron microscopy of an aggregate of astrovirus VA1 particles in a Caco-2 cell 
Infection of mouse hearts with Astrovirus VA1 (green), nuclei labeled with DAPI (blue) 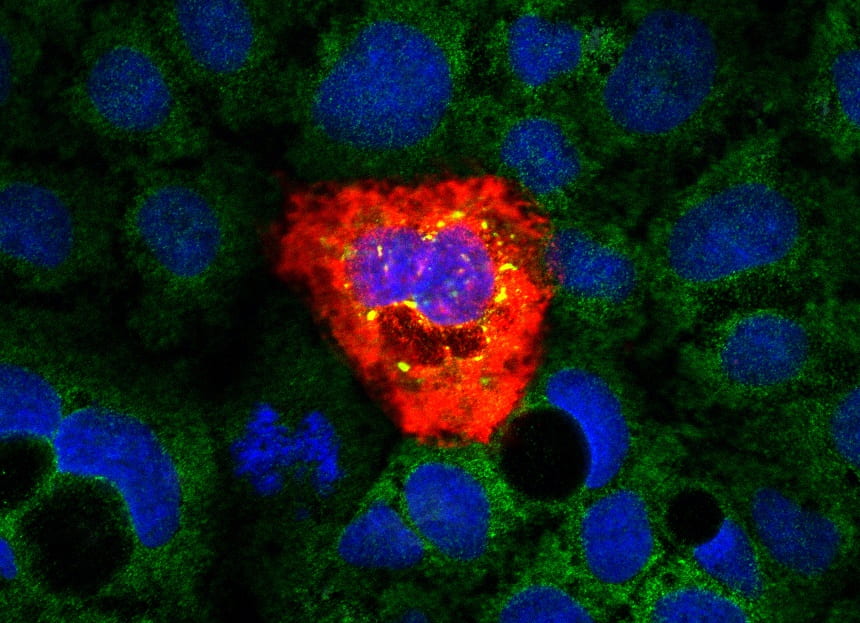
Immunofluorescence of a Caco-2 cell infected with astrovirus VA1 (red). eIF3A is stained in green. Stress granules containing eIF3A are induced upon infection (yellow). Nuclei stained with DAPI (blue)